Biosignal processing for automatic emotion recognition
By Danielle Benesch, & Achraf Kabbabi
Published on June 12, 2020
June 12, 2020

Project definition
Background
About us
Danielle
I started my research master’s this January, and my thesis project will involve the automatic classification of biosignals among other things. My background is in cognitive science, and while I’ve already learned about the basics of machine learning and neuroinformatics, I would like to improve my skills by working on a practical project with real data.
Achraf
I am currently studying Biomedical Engineering at Polytechnique Montréal (M.Eng) and am enrolled in the Brain Hack School 2020 adventure for growing my technical skills as well as networking. I have been modestly introduced to NeuroImaging before enrolling in the biomedical engineering path and would like to broaden my knowledge of the field and sharpen my skills in it.
About the project
Psychological stress has been found to be associated with changes in certain biosignals. Features extracted from these biosignals have increasingly been used for predicting an individual’s emotional state, including the EEG alpha asymmetry index, heart rate variability, and skin conductance response (Giannakakis et al., 2019). This research has allowed for the development of technologies that can respond to users’ emotions (Carroll et al., 2013) and motivated the creation of multiple datasets containing emotion-correlated biosignal data (Wiem & Lachiri, 2017). In this project, we were broadly interested in the task of emotion recognition, using a publicly available dataset to explore how we could predict emotions using biosignals.
Tools
Tools and techniques we used:
- Bash and Python


- Git and GitHub

- Preprocessing and feature extraction with Scipy and Neurokit


- Data visualization with Plotly

- Machine learning with Scikit-learn

Data
At the start of the project, we considered using the following databases:
- DREAMER: A Database for Emotion Recognition through EEG and ECG Signals from Wireless Low-cost Off-the-Shelf Devices
- Biosignal data: EEG and ECG
- Emotion data: Rating on valence-arousal-dominance scale provided by participants and tags for the “target emotion” of the stimuli
- The MAHNOB-HCI-Tagging database
- Biosignal data: EEG (in the form of BDF files), ECG, respiration amplitude, and skin temperature
- Emotion data: Rating on valence-arousal scale provided by participants, and for some of the data, emotion tags (e.g. amused) selected by participants
- An EEG dataset recorded during affective music listening
- Biosignal data: EEG
- Emotion data: Rating on valence-arousal scale provided by participants (though it’s not immediately clear where this can be downloaded)
- AffectiveROAD system and database to assess driver’s attention.
- Biosignal data: BVP, EDA, ECG, respiration rate, skin temperature
- Emotion data: “Stress metric” provided by observing experimenter
Ultimately, we decided to use the DREAMER dataset which must be requested from the authors here.
The dataset contains EEG and ECG data from 23 participants were shown 18 videos intended to elicit 9 different emotions - as well as “neutral videos” thought to have no valence for “baseline” data. Biosignal data was collected using the Emotiv EPOC wireless EEG headset and the Shimmer2 ECG sensor. We were especially interested in how accurately we could classify emotions using biosignal data collected by portable, inexpensive devices due to the potential of automatic emotion recognition incorporated into wearables. An image of the equipment is shown below (from Katsigiannis & Ramzan, 2018).

More information on how the data were collected can be found in the PDF DREAMER_info (downloaded from Zenodo) or in the paper by Katsigiannis and Ramzan (2018).
Deliverables
We were able to complete:
Data preprocessing and feature extraction for at least one biosignal.
Visualization of the relationship between the extracted features and the emotion data.
Both interactive and static visualizations
Training a classifier to predict the emotion data.
More information about the contents of this repository and instructions for how to run them can be found in the “repoInfo” Markdown file located in the docs folder.
The project presentation is available here.
Progress & Results
We first preprocessed the biosignals and explored the relationship of extracted features with the emotion data, then we evaluated the performance of a number of classifiers. We created a minimal python script that performs the data preprocessing, feature extraction, and the classifier evaluation.
Achraf: Preprocessing and feature extraction
The preprocessing scripts I wrote were inspired by Jiaqi1008’s repository. The DREAMER dataset being a .mat file, I used the library Scipy to load it: it contained EEG data, ECG data, and subjective ratings. The preprocessing for EEG data consisted of extracting the maximum of the Power Spectrum Density (PSD) for the EEG signals for three bands (theta, alpha, beta), for each of the 14 electrodes used. The library Scipy was used for filtering and PSD extraction (Welch’s method). The preprocessing for ECG data was done thanks to the library Neurokit2 by first preprocessing the data with the ecg_process() method then by extracting the features with the ecg_intervalrelated() method. The features extracted were the Mean Heart Rate and various Heart Rate Variability (HRV) metrics. I tested those preprocessing pipelines in notebooks first, then I wrote a script DREAMER_main.py implementing them. An output example of the script in a terminal can be found here
Danielle: Evaluation of classifiers
I was interested in training a classifier to detect stress, but at the beginning of the project, I wasn’t sure how exactly we should use the data to do so. I wanted to be able to compare my results to others, but there appears to be a wide range of ways to evaluate stress detection, further complicated by the fact that research on automatic emotion recognition and stress detection has relied on different models of emotion (Wiem & Lachiri, 2017).
We first considered predicting self-reported valence and arousal scores, but then we explored the data and looked at the relationship between some of the features we extracted and valence/arousal. It seemed like these features did not indicate changes in valence or arousal as much as we’d hoped. It would be interesting to see how well we could predict valence and arousal using different methods of preprocessing, feature extraction, scaling, thresholding etc., but for the sake of this project, we thought we’d start with an easier classification problem.
In the paper describing DREAMER (Katsigiannis & Ramzan, 2018), Table I contains the “target emotion” of each video clip with the video ID. The target emotion was not included in the .mat file downloaded from Zenodo, so I added this information to each datapoint based on the video ID. Including this information allowed us to predict the emotions that were intended to be evoked by each stimulus, rather than self-reported emotions.
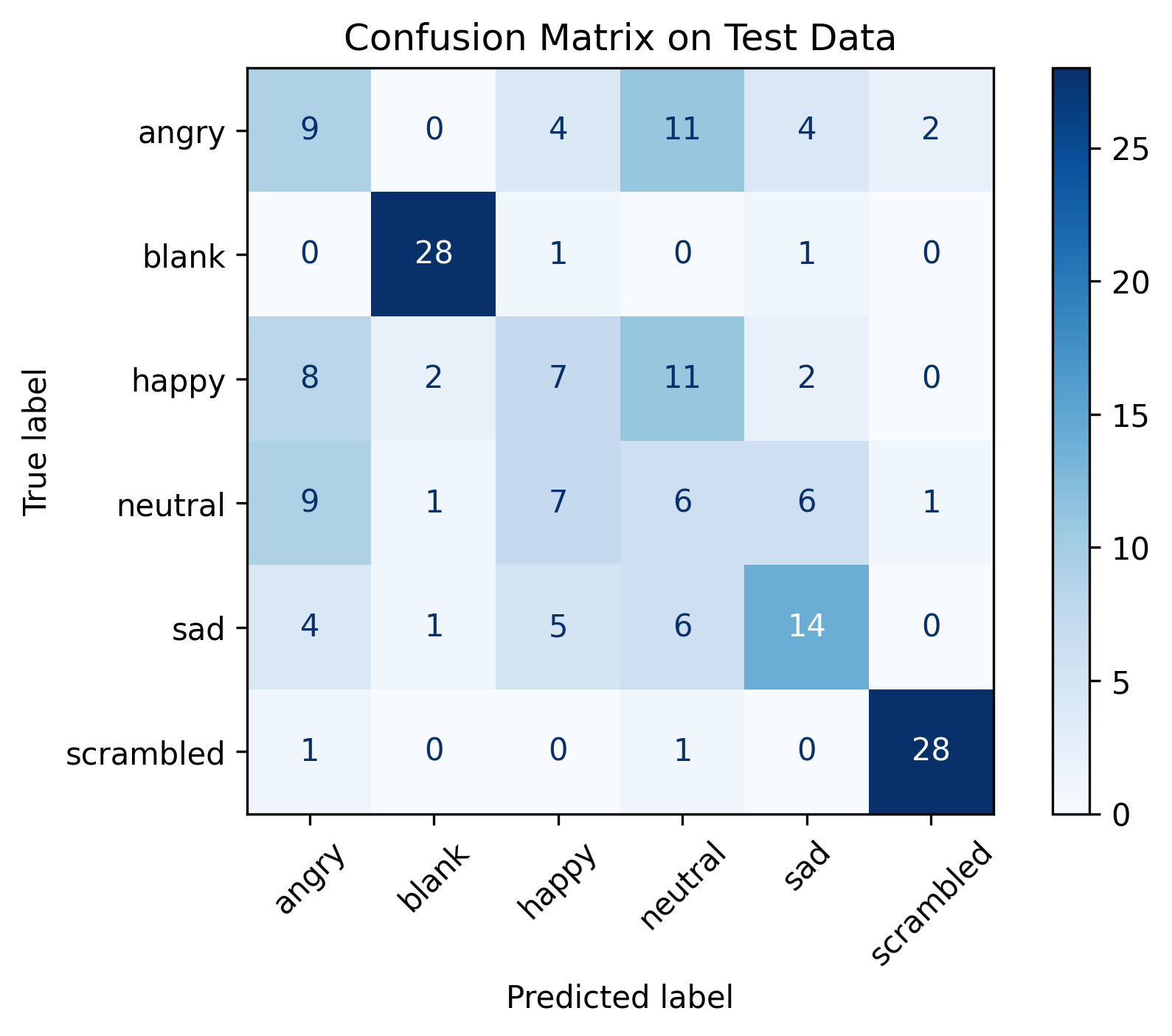
There were 9 different target emotions: “calmness”, “surprise”, “amusement”, “fear”, “excitement”, “disgust”, “happiness”, “anger”, and “sadness”. Before determining whether we could predict the full sprectrum of emotions, I wanted to see whether we could distinguish “calmness” from two emotions on the opposite side of the spectrum: “anger” and “fear”. This constituted a binary classification task: “calmness” vs. “not calmness”.
To try to get an idea of how the classifiers would perform given biosignal data from completely new people, I evaluated the classifiers using Group 10-Fold Cross-Validation, with the groups being the participants - meaning that the training set and validation set always consisted of data from separate participants. However, we should note that we used the entire dataset for data exploration; there was no held-out test set that we only used solely for evaluation. Evaluating the classifiers on another emotion-correlated biosignal dataset may provide a more realistic idea of the classifiers’ generalizability.
I selected a number of classifiers based on a script from the sci-kit learn documentation: Nearest Neighbors, Support Vector Machine with a linear kernel, Support Vector Machine with an RBF kernel, Gaussian Process, Decision Tree, Random Forest, Multi-layer Perceptron, AdaBoost, and Naive Bayes. The accuracy averaged over the splits for each classifier, with features extracted from both the EEG and ECG data, is shown below:
| Name | Mean Accuracy |
|---|---|
| Nearest Neighbors | 0.87 |
| Linear SVM | 0.67 |
| RBF SVM | 0.87 |
| Gaussian Process | 0.7 |
| Decision Tree | 0.9 |
| Random Forest | 0.9 |
| Neural Net | 0.73 |
| AdaBoost | 0.98 |
| Naive Bayes | 0.59 |
AdaBoost, an ensemble method, had the highest mean accuracy. I also measured the average prediction runtime, and AdaBoost was relatively slow compared to the other classifiers (about 5-10 ms, while most of the others were around 1 ms, on my machine). While we didn’t get to do this during this project, it would be interesting to see how methods to reduce runtime affect performance.
Week 3 deliverable: data visualization
Danielle’s Week 3 deliverables
- Interactive Figure: “Participant Ratings of Film Clips in Valence-Arousal Space
- Bonus: a jupyter notebook generating this figure… written while using a linter, which I know about thanks to Greg’s talk on scripting!
- Bonus: a jupyter notebook generating this figure… written while using a linter, which I know about thanks to Greg’s talk on scripting!
- Interactive Figure: “Group 10-Fold Cross Validation with DREAMER data”
- Interactive Figure: “Score vs. Prediction Runtime for all CV Iterations and Classifiers”
Achraf’s Week 3 deliverables
- A Jupyter Notebook generating static and interactive visuals (ipynb) and the corresponding HTML version
- Static visuals that I found interesting during visual exploration of the data:
- Screenshot of terminal outputs of the scripts we wrote for Compute Canada (minimal version):
Conclusion and acknowledgements
During the BrainHack School, we hoped to learn as much as we could about modern (neuro)scientific practice, to improve our Python skills through a hands-on multi-disciplinary project, and to exchange information and expertise with the other participants.
The first week was an intense theoretical and practical overview of many modern tools in neuroimaging, many of which we had very limited experience with or had never even heard of. The second week taught us how important it is to take enough time to clearly define a project and specify goals for it. Starting with a relatively broad project, it was difficult to narrow our goals down to one dataset and method, but ultimately we chose to pick one and see it through to the end.
The third and fourth weeks were the core of the brain-hacking adventure and taught us many things such as:
- Improved coding skills: Python notebooks & scripting
- Deeper understanding of machine learning
- Open science tools and practices
While we weren’t able to incorporate all of the tools that we had hoped to use in our project, we are grateful to have been exposed to them. Knowing where we can find helpful online resources on all of the topics covered, we hope to slowly integrate what we’ve learned into our own work, post-BrainHack.
We would like to thank the course organizers and our instructors who spent a lot of time helping us with of our project and generally enlightening us:
- Greg
- Agâh
- Loic
- Yann
References
Carroll, E. A., Czerwinski, M., Roseway, A., Kapoor, A., Johns, P., Rowan, K., & Schraefel, M. C. (2013, September). Food and mood: Just-in-time support for emotional eating. In 2013 Humaine Association Conference on Affective Computing and Intelligent Interaction (pp. 252-257). IEEE.
Giannakakis, G., Grigoriadis, D., Giannakaki, K., Simantiraki, O., Roniotis, A., & Tsiknakis, M. (2019). Review on psychological stress detection using biosignals. IEEE Transactions on Affective Computing.
Katsigiannis, S., & Ramzan, N. (2017). DREAMER: A database for emotion recognition through EEG and ECG signals from wireless low-cost off-the-shelf devices. IEEE journal of biomedical and health informatics, 22(1), 98-107.
Wiem, M. B. H., & Lachiri, Z. (2017). Emotion classification in arousal valence model using MAHNOB-HCI database. Int. J. Adv. Comput. Sci. Appl. IJACSA, 8(3).