Decoding Depression via EEG Biomarkers: A Neurocomputational Approach using Machine and Deep Learning
By Wei-Xuan Chai
Published on June 15, 2025
June 15, 2025

Project definition
Background
This project was inspired by a clinical case from Shanghai Ruijin Hospital, where Brain-Computer Interface (BCI) technology was applied to treat severe depression. That real case made me wonder about the potential of neural decoding in understanding and supporting mental health.
As someone interested in both neuroscience and BCI, I believe it is essential to master machine learning and deep learning techniques—especially when working with high-dimensional signals like EEG or other neuroimaging data. This project is my personal attempt to explore how machine learning (SVM) and deep learning (EEGNet) models perform in classifying major depressive disorder (MDD) based on EEG band power features.
Tools
The project will rely on the following technologies:
- Programming & Environment
- Python
- Jupyter Notebook: Used for data preprocessing and training the SVM machine learning model.
- Google Colab: Used for training the EEGNet deep learning model.
- Libraries & Frameworks
- scikit-learn, gridsearchCV: For machine learning models.
- MNE: For EEG data processing and visualization.
- numpy, pandas, matplotlib: For data manipulation and plotting.
- tensorflow, keras: For deep learning models.
- External Models / Resources
- Brainhack School Modules: Working with MNE-Python and EEG-BIDS page
- Brainhack School Modules: Machine learning for neuroimaging page
- EEGNet implementation from ARL (Army Research Laboratory) EEGModels Project page
Data
This project uses the MDD Patients and Healthy Controls EEG Data (New), downloaded from page.
- The dataset includes 30 Healthy Controls (H) and 34 Major Depressive Disorder (MDD) participants.
- Each participant completed three conditions:
- Eyes Closed (EC)
- Eyes Open (EO)
- P300 Task (TASK)
For this analysis, we used:
- Only the 5-minute EC EEG data
- Successfully downloaded: 28 H and 30 MDD subjects
Deliverables
At the end of this project, we will have:
- Preprocessing & Bandpower Feature.ipynb
- Machine Learning_SVM.ipynb
- Deep Learning_EEGNet.ipynb These deliverables reflect the full pipeline from raw EEG preprocessing to both traditional ML and DL models training.
Results
Progress overview
This project started with the goal of integrating what I’ve previously learned—EEG preprocessing, machine learning, and deep learning—into a single end-to-end pipeline. While I had prior experience working with EEG data and individual modeling techniques, I had never tried building an entire classification workflow on my own, from data preparation to model comparison.
I began by preprocessing the EEG data using MNE-Python and extracting temporal-domain features (band power). Then I implemented an SVM classifier, followed by a deep learning model based on EEGNet. Although the EEGNet part was more challenging due to model tuning and GPU limitations, I still managed to get a working prototype on Google Colab.
Beyond modeling, I also learned how to structure a project repository, push updates to GitHub, and write clear documentation—sometimes after a few frustrating errors (and moments of panic). But overall, it was a fun and fulfilling learning experience, and a solid first attempt at decoding mental health signals using both machine learning and deep learning techniques.
Tools I learned during this project
- Learned to preprocess EEG signals and extract bandpower features using MNE.
- Applied classical machine learning models (SVM) with scikit-learn and gridsearchCV.
- Implemented and trained a deep learning EEG model (EEGNet) with TensorFlow and Keras.
- Developing, testing, and executing the entire analysis pipeline using Jupyter Notebooks and Google Colab as interactive environments
Results
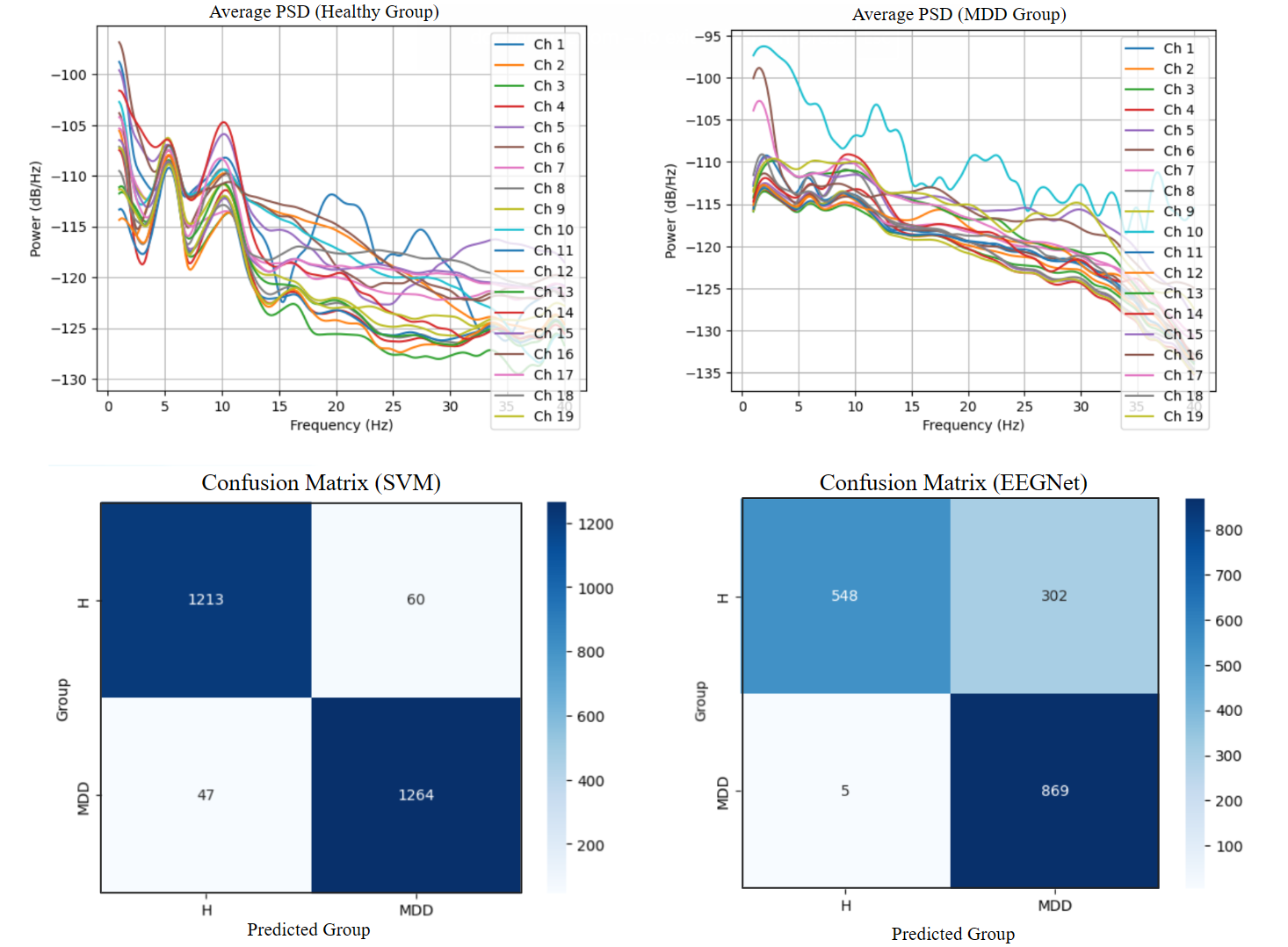
Preliminary results suggested that SVM achieved higher accuracy (> 0.95) compared to EEGNet (~0.8), possibly due to limited hyperparameter tuning and model optimization in this early-stage exploration. While deep learning provides promising end-to-end modeling capabilities, classical approaches like SVM may still offer superior performance when the dataset is small and features are carefully extracted.
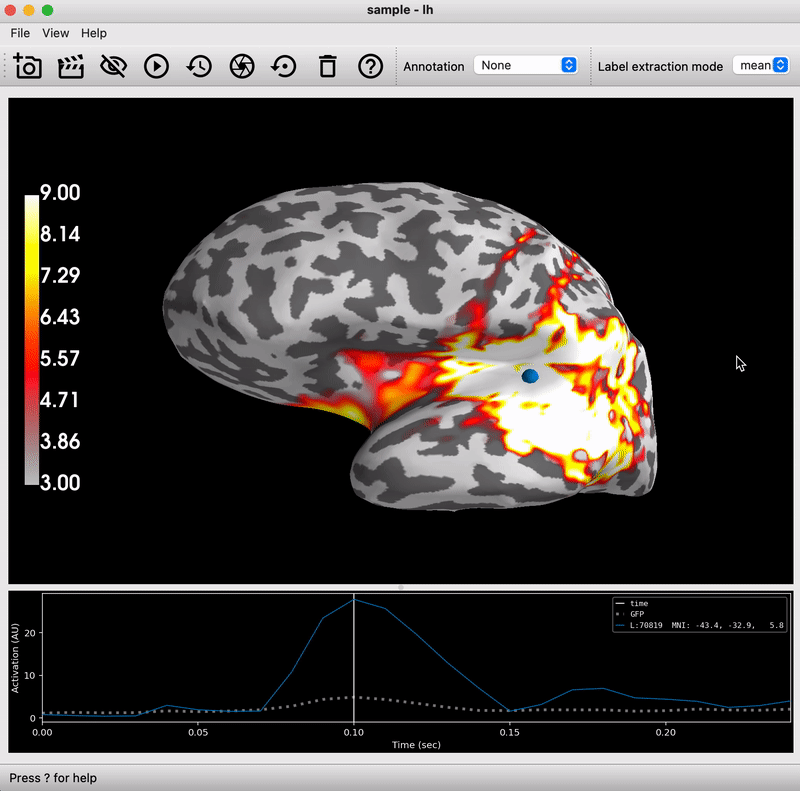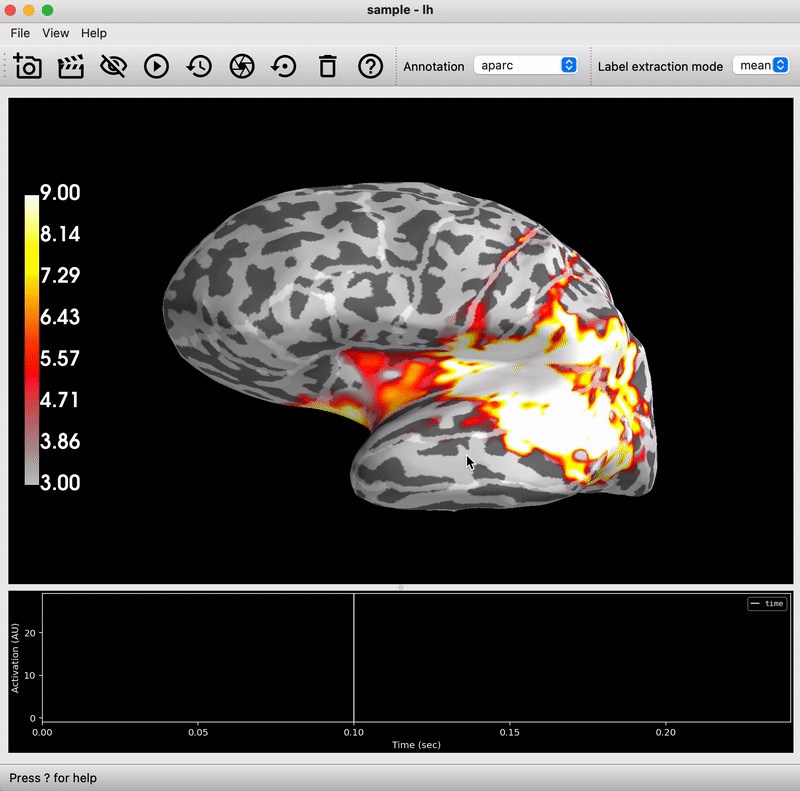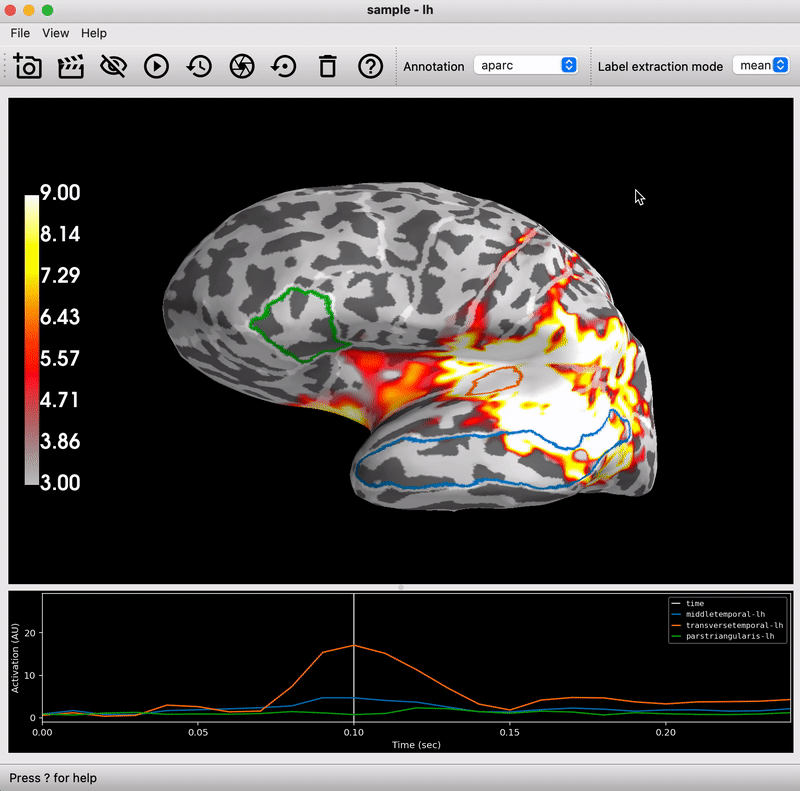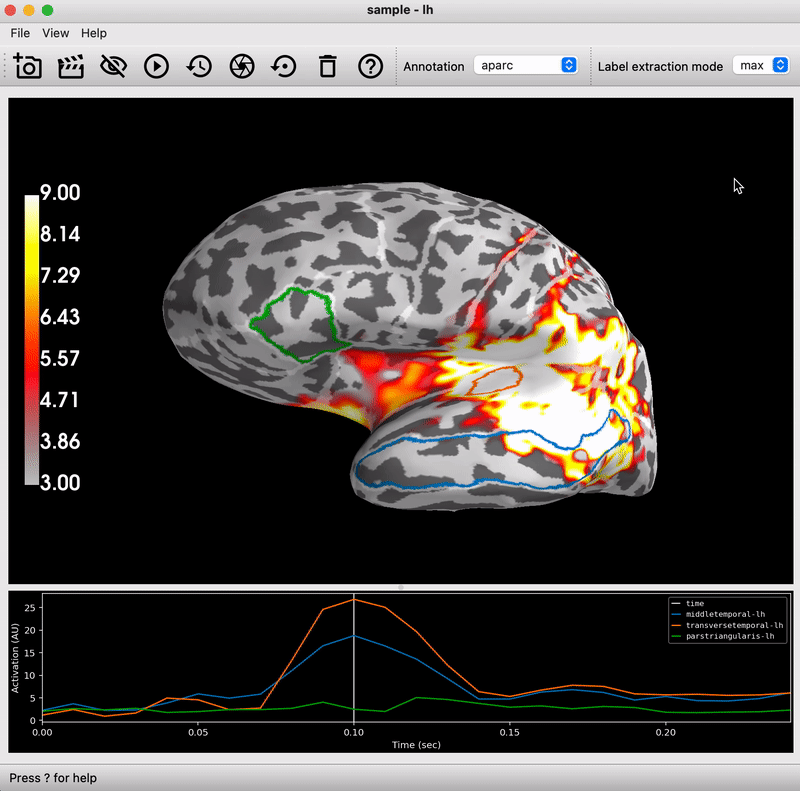
Deliverable 1: Preprocessing & Bandpower Feature
An EEG preprocessing pipeline was implemented using MNE-Python, followed by the extraction of frequency-domain features (band power) for classification purposes.
Deliverable 2: Machine Learning_SVM
A Support Vector Machine (SVM) model was trained and evaluated based on the extracted band power features. Grid search with 10-fold cross-validation was used to tune key hyperparameters (e.g., C, gamma). Several trials were conducted, including the integration of feature selection and dimensionality reduction using PCA. However, these modifications did not improve the accuracy, suggesting that the original band power features already effectively captured discriminative patterns related to depression in the EEG data, or that such techniques may not be beneficial in this particular case.
Deliverable 3: Deep Learning_EEGNet
An EEGNet model page was trained and evaluated using preprocessed EEG epoch data, without extracting band power features. The model was implemented on Google Colab with GPU support. Hyperparameter tuning was conducted using both Keras Tuner and manual for-loop implementations, each within a 10-fold cross-validation framework. Due to time constraints, only minimal tuning was performed, which may explain the lower performance (~80% accuracy). Nevertheless, this pipeline demonstrates how CNN-based architectures can be applied end-to-end to EEG data.
Conclusion and acknowledgement
This project was my first attempt at an EEG decoding pipeline, from preprocessing, feature extraction, to model training with SVM and EEGNet. Though basic, it revealed skill gaps in computational neuroscience and BCI, guiding my future learning.
I appreciate the supportive learning environment and resources from Brainhack School 2025, including video tutorials and coding notebooks. Special thanks to the researchers who shared the MDD EEG dataset on Figshare and the EEGNet developers for their open-source contribution. This project reflects the power of open science and community collaboration.