CVAE-based ADHD neuroimaging analysis
By Cian-Ya Lan, & Jia-Ling Sun
Published on June 13, 2025
June 13, 2025

Project definition
Background
- Reconstruct 3D brain MRIs using CVAEs.
- Disentangle “salient” ADHD-related features from “background” features common to both ADHD and typically developing children.
- Assess how well the learned latent spaces reflect behavioral and clinical variation using:
- Silhouette Analysis
- Representational Similarity Analysis (RSA)
Tools
This project used:
- Python (NumPy, Pandas, SciPy, Scikit-learn, Matplotlib, Seaborn)
- Keras (TensorFlow backend) for deep learning
- UMAP for latent space visualization
- Representational Similarity Analysis (RSA) using Kendall’s tau
- Silhouette analysis for latent space separability
- GitHub for version control
Data
This project used the publicly available ADHD-200 dataset, specifically the Burner-preprocessed version:
- Structural MRI data processed via voxel-based morphometry (VBM)
- Normalized 3D gray matter volumes (64×64×64)
- Accompanied by phenotypic variables: age, sex, diagnosis, subtype, medication, IQ, etc.
Deliverables
At the end of the project, we produced:
- A working CVAE framework for modeling neuroanatomical variation
- Visualizations of latent features and synthetic brain reconstructions
- RSA and clustering results relating brain features to clinical data
- Well-documented code and reproducible analysis notebooks
Results
Progress overview
We trained a CVAE model with two latent components:
s(salient features): ADHD-specific anatomical variationz(background features): common/shared variation
We evaluated the model using:
- Silhouette scores for latent clustering
- RSA to correlate latent dimensions with phenotypic variables
- GMM clustering and BIC to test for discrete vs. continuous subtype structure
Tools I learned during this project
- Contrastive representation learning in generative models
- Implementation of CVAEs in Keras
- Preprocessing and working with VBM MRI data
- Representational Similarity Analysis
- Model interpretability techniques for brain data
Results
Deliverable 1: CVAE analysis
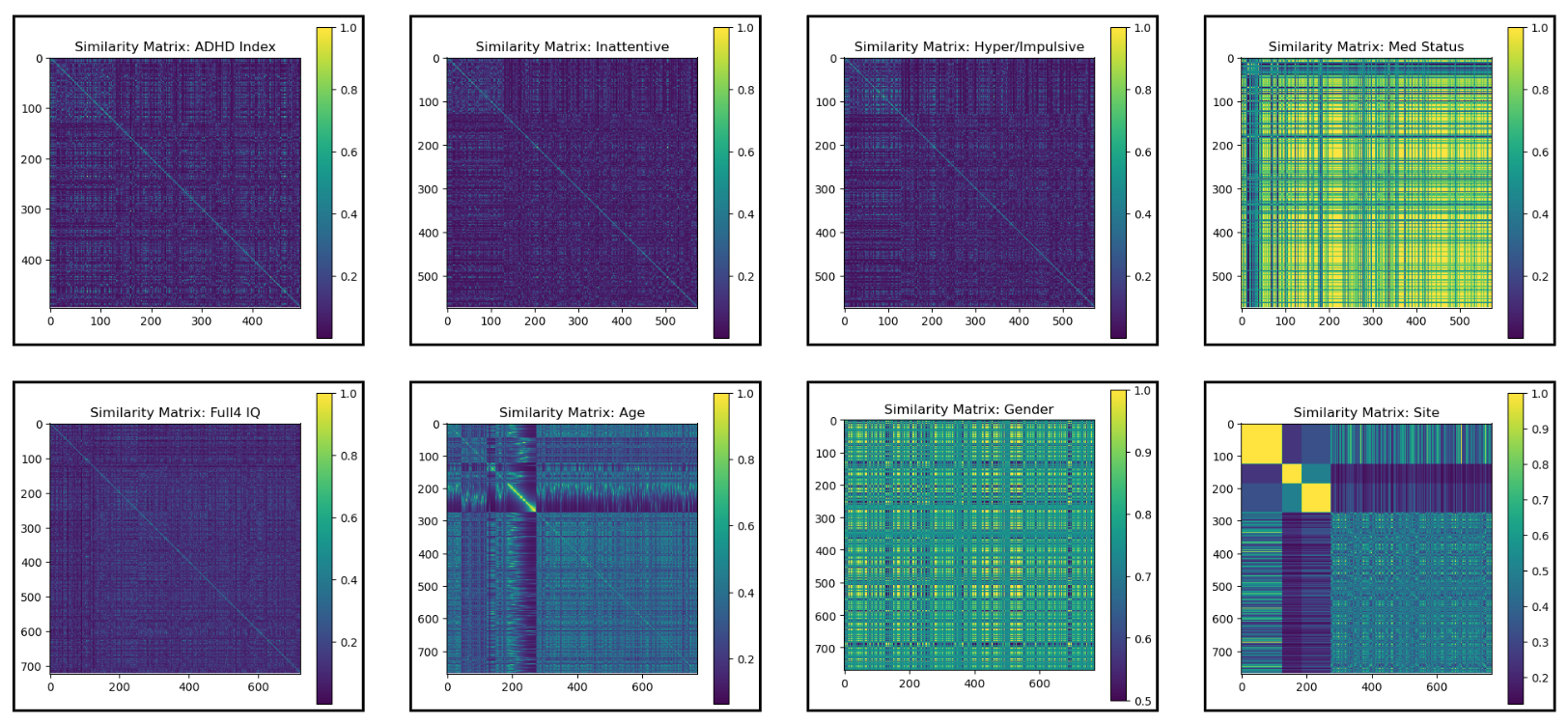
Salient features (
s) were significantly correlated with:- ADHD Index
- Inattentive and Hyperactive/Impulsive scores
- Medication status and age
Shared features (
z) were more related to:- IQ, gender, and scan site
Deliverable 2: Clustering analysis
- Gaussian Mixture Models (GMM) with Bayesian Information Criterion (BIC) showed:
- Lowest BIC at 1 cluster, suggesting continuous heterogeneity
- Results align with dimensional models of psychiatric disorders
Deliverable 3: Code and notebook
- Full training pipeline in
Train-CVAE-ADHD200.ipynb - Visualization and RSA in helper scripts
- Readme documentation and reproducibility checklist included
Conclusion and acknowledgement
This project demonstrates the potential of contrastive deep generative models like CVAEs to disentangle disorder-specific neuroanatomical features from shared variation. Our findings suggest ADHD may be better described along a spectrum rather than discrete subtypes. We thank the Brainhack School instructors and the open neuroimaging community for providing tools and data that made this project possible.