Increasing Accessibility of BrainAGE Open Access Calculators
By Jiya Shah, Eleanor Pak Wai Lam, Isabella Monaghan Chow, & Khubaib Choudry
Published on June 19, 2025
June 19, 2025

Project definition
Background
Our group
We are a group of MSc students from the Institute of Medical Science at the University of Toronto, conducting our thesis research in collaboration with the Centre for Addiction and Mental Health (CAMH). Although our individual projects examine diverse aspects of mental health through various methodologies, we share a strong collective interest in advancing research in this field.
Project background
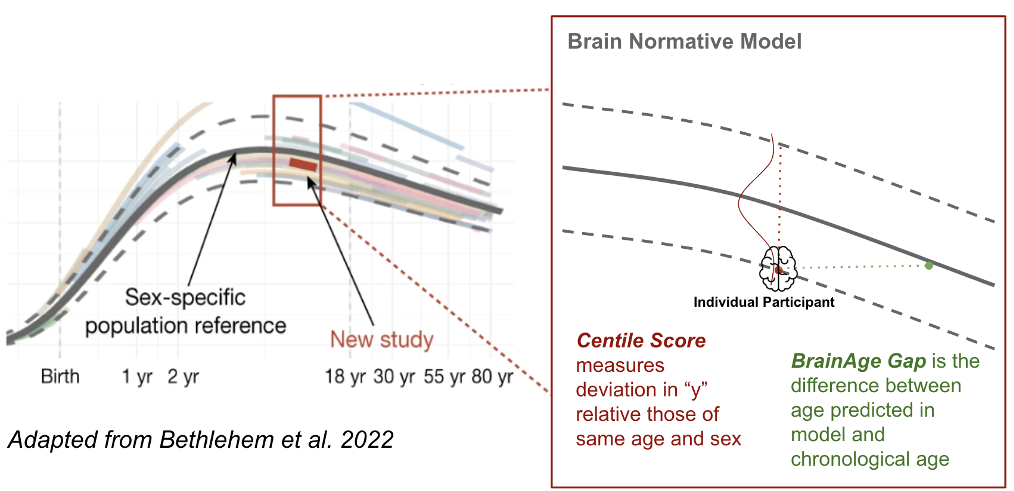
Normative brain age modelling is a technique that uses structural MRI (sMRI) data to estimate an individual’s brain age by comparing it to a normative reference group. The difference between predicted brain age and chronological age, known as the Brain Age Gap Estimate (BrainAGE),can provide insights into atypical brain aging or neurodegenerative risk. A number of open-access BrainAGE calculators have been developed, but they often differ in required input features, preprocessing steps, and anatomical atlases, making them difficult to use without specialized technical knowledge. This lack of standardization and unclear input requirements pose barriers to accessibility and reproducibility across studies.
Tools
The project utilizes a range of tools and platforms to support development, reproducibility, and open dissemination:
- R Programming Language: Core environment for data processing and script development.
- RMarkdown: Integrates code, analysis, and narrative into reproducible documents.
- Git & GitHub: Enables version control, collaborative development, and public access to project materials.
- Visual Studio Code (VS Code): Integrated development environment for managing project files and workflows.
- Targeted BrainAGE Models: Input standardization tools were developed for a variety of open-access calculators, including: Whole-brain models: NAPR, Developmental BrainAGE, Kauffman BrainAGE, and BrainAGE Region-/tissue-specific models: CentileBrain, Network-BrainAGE, and BrainChart.io
Data
This project uses normative neuroimaging data from the Reproducible Brain Charts initiative, specifically the Healthy Brain Network (HBN) subset. The HBN provides openly available, preprocessed structural MRI features from a large developmental cohort (ages 5–21).
Deliverables
Our main deliverable is a publicly accessible GitHub repository housing all RMarkdown tools and documentation for standardizing BrainAGE calculator inputs.
Results
Progress overview
The final presentation of this project was delivered on May 30, 2025. All planned deliverables were completed and are available in the project’s GitHub repository. All scripts have been tested with the HBN dataset and are running successfully. Initial issues related to column matching and output formatting have been resolved.
Tools we learned during this project
- Git & GitHub: Collaborating on RMarkdown scripts and resolving issues highlighted the critical role of Git. We gained hands-on experience using Git for version control and GitHub for team collaboration, particularly in troubleshooting and merging code.
- Bash/Terminal: We developed proficiency in fundamental command-line operations (e.g., cd, ls, mkdir) for navigating directories, managing files, and organizing project outputs, which are skills essential for handling data and executing scripts efficiently.
- R Programming: SFor some team members, this project was their first opportunity to use R extensively. It enabled us to learn and build skills in data wrangling, scripting, and applying R for more complex data manipulation and analysis within the project’s scope.
Results
Deliverable: A GitHub Repository
We developed a suite of RMarkdown scripts to standardize and streamline data preparation for open-access BrainAGE calculators. These tools are publicly available in a centralized GitHub repository (github.com/shahjiya/brainhack_brainage_project), providing researchers with easy access to reproducible workflows and comprehensive documentation.
Key contributions include:
HBN RBC Data Preparation Guide: An RMarkdown document (HBN_data.Rmd) that presents a reproducible pipeline for downloading structural MRI data from the Healthy Brain Network Reproducible Brain Charts (HBN RBC) dataset, aggregating files into CSV format, and reformatting regional surface statistics to ensure compatibility with multiple brain atlases and BrainAGE models.
Calculator-Specific Data Wrangling Scripts: Seven RMarkdown scripts designed to transform demographic and MRI data into the precise input formats required by various BrainAGE calculators. These scripts automate column mapping, renaming, type conversions, and output generation to facilitate seamless integration. The scripts are: BrainAGE_regular.Rmd, Developmental_brain_age.Rmd, NAPR_analysis.Rmd, Network_BrainAGE.Rmd, brainchart.io.Rmd, centilebrain_template.Rmd, kauffman_brainage.Rmd
Conclusion
This project lays the foundation for improving the accessibility and reproducibility of open-access BrainAGE calculators by standardizing input formats and simplifying data preparation. The tools can be adapted to other datasets or extended to additional BrainAGE models. Future work may include developing a web-based interface to further reduce technical barriers and expand the use of brain age modelling in neuroimaging research.
Acknowledgement
We would like to thank BrainHack School 2025 for organizing this course and creating a supportive learning environment. As a team with diverse coding experience, we faced many challenges along the way. We are especially grateful to our group members, course instructor, and TA for their patience, guidance, and encouragement throughout the project.
References
- Franke K, Ziegler G, Klöppel S, Gaser C. 2010. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. NeuroImage. 50(3):883–892. https://doi.org/10.1016/j.neuroimage.2010.01.005
- Cole JH, Ritchie SJ, Bastin ME, Valdés Hernández MC, Muñoz Maniega S, Royle N, Corley J, Pattie A, Harris SE, Zhang Q, et al. 2018. Brain age predicts mortality. Mol Psychiatry. 23(5):1385–1392. https://doi.org/10.1038/mp.2017.62
- Rokicki J, Wolfers T, Nordhøy W, Tesli N, Quintana DS, Alnæs D, Richard G, Lange AM, Lund MJ, Norbom LB, et al. 2021. Multimodal imaging improves brain age prediction and reveals distinct abnormalities in patients with psychiatric and neurological disorders. Hum Brain Mapp. 42(6):1714–1726. https://doi.org/10.1002/hbm.25323
- Habes M, Pomponio R, Shou H, Doshi J, Mamourian E, Erus G, Nasrallah IM, Launer LJ, Rashid T, Bilgel M, et al. 2021. The Brain Chart of Aging: Machine-learning analytics reveals links between brain aging, white matter disease, amyloid burden, and cognition in the iSTAGING consortium. Alzheimers Dement. 17(1):89–102. https://doi.org/10.1002/alz.12178
- Boyle R, Knight SP, de Lange AMG, Abe Y, Kievit RA. 2021. Brain-predicted age difference score is related to specific cognitive functions: a multi-site replication analysis. Brain Imaging Behav. 15(1):327–345. https://doi.org/10.1007/s11682-020-00321-9