Multimodal spine multiple sclerosis segmentation
By Thomas Dagonneau
Published on May 16, 2025
May 16, 2025

Project definition
Background
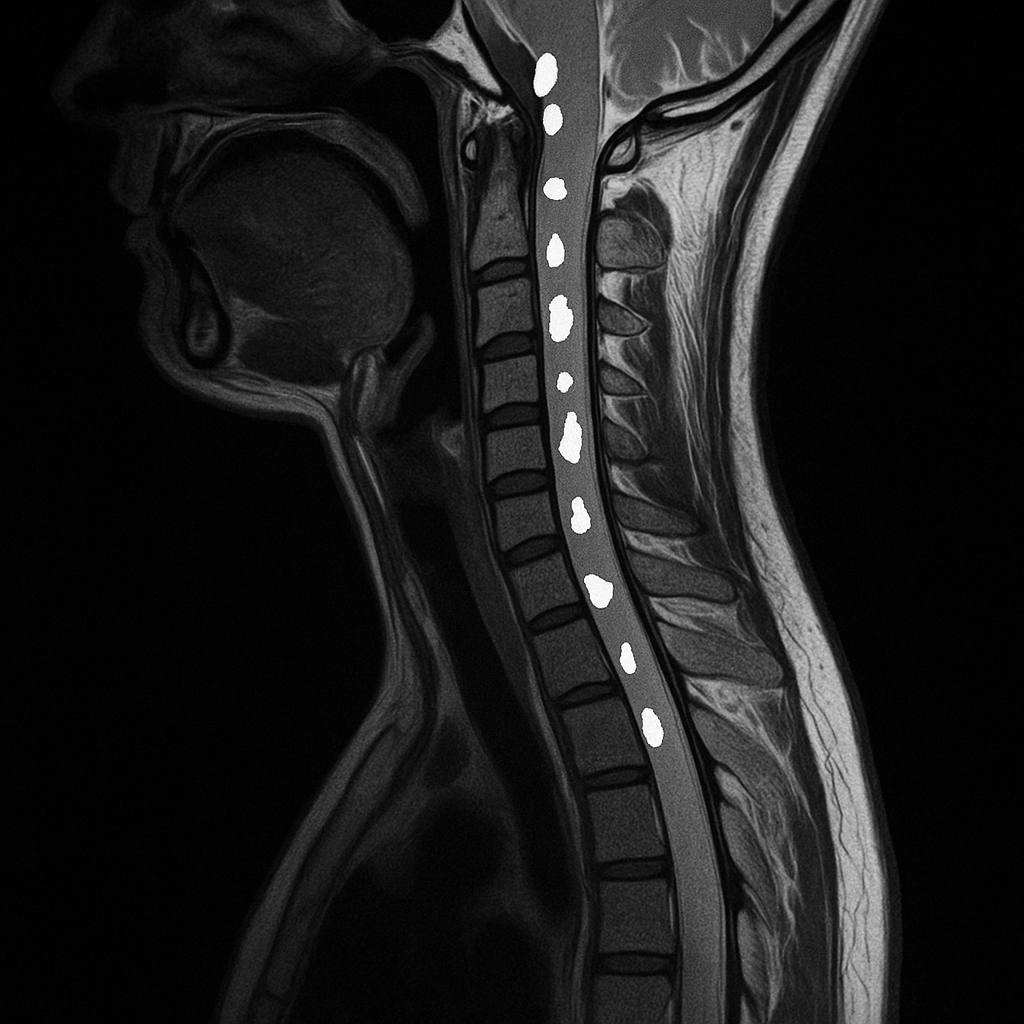
Multiple sclerosis (MS) is a chronic disease of the central nervous system, with spinal cord involvement playing a critical role in clinical outcomes. Although MRI is essential for MS diagnosis and disease monitoring [1], spinal cord imaging remains challenging due to anatomical complexity and variability in acquisition protocols. Existing automated lesion segmentation tools have focused primarily on the brain [2–4], and very few methods were designed for spinal cord lesions, particularly across heterogeneous and multimodal MRI data. Additionally, high inter- and intra-rater variability in manual annotations [5] highlights the need for reliable, automated segmentation pipelines.
In this study, we introduce a novel pipeline for the automatic segmentation of MS lesions in the spinal cord using multiple MRI acquisitions from the same patient. Our approach combines a deep learning model with a robust post-processing framework to produce accurate lesion instance segmentations and estimate prediction uncertainty, addressing the specific challenges of spinal cord imaging.
Objectives
In this project, I focus on the Multiple Sclerosis Spinal Cord Lesions Detection from MultiSequence MRIs Challenge (MS-Multi-Spine) and develop a model capable of multimodal MS lesion segmentation.
- Train a multimodal nnUNet [6] on the challenge’s dataset (private)
- Make the model and pipeline open-source on GitHub (public)
- Containerize the pipeline and provide a Boutiques descriptor to facilitate reuse (public)
Tools
This project uses the following tools:
- Terminal: all code is designed to run in the terminal
- Python: all scripts are written in Python
- Spinal Cord Toolbox (SCT) [7]: used for preprocessing steps such as coregistration
- Git and GitHub: for version control and open access
- Docker: to ensure reproducibility
- Boutiques: to automate and simplify tool usage
Data
The dataset used in this project is provided by the challenge and is private. It includes:
- 100 subjects in total
- 50 subjects with T2w + STIR acquisitions
- 25 subjects with T2w + PSIR acquisitions
- 25 subjects with T2w + MP2RAGE acquisitions
Deliverables in Week 4
- A GitHub repository containing Python scripts for data preprocessing, inference, and postprocessing
- A Dockerfile used to create the Docker image available here
- A Boutiques descriptor to perform inference
Results
Over the four weeks, I developed a pipeline for the automatic segmentation of MS lesions in a multimodal context. I gained hands-on experience with:
- Python scripting: all processing steps are implemented in Python
- Git and GitHub: all project code and resources are versioned and publicly available
- Spinal Cord Toolbox: used for preprocessing (e.g., coregistration)
- Docker: used to package the pipeline and publish the image to Docker Hub
- Boutiques: used to create a descriptor for automated execution of the tool
Deliverable 1: GitHub Repository
This repository contains everything needed to reproduce the training, inference, and submission process. Instructions to train and perform inference are available here.
Deliverable 2: Docker
The repository includes the Dockerfile used to build the image available on Docker Hub. Instructions for building the image locally are provided here.
Deliverable 3: Boutiques Descriptor
The Boutiques descriptor is available here, with usage instructions here.
Contribution to Open Science
This project contributes to open science by:
- Publishing the complete pipeline code and documentation on GitHub
- Providing a Docker image to ensure reproducibility across environments
- Creating a Boutiques descriptor to facilitate automated reuse by other researchers
- Building on and extending open-source tools such as nnUNet and SCT
- Enabling multimodal spinal cord lesion segmentation—an underexplored area in neuroimaging research
Conclusion
This project presents a complete pipeline for multimodal MS lesion segmentation in the spinal cord, from preprocessing to inference and postprocessing. It leverages state-of-the-art tools and open standards to enable reproducibility and accessibility. The pipeline successfully integrates multimodal imaging data and handles the complexity of spinal cord anatomy, providing reliable segmentation outputs.
Next Steps
- Evaluate the model on additional test data and quantify performance per modality
- Extend support to include missing modalities through imputation or modality-agnostic models
- Release a demo dataset to allow users to test the pipeline without requiring private data
- Submit the pipeline to MICCAI MS Challenge 2025 results and benchmark against other approaches
References
- Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17: 162–173.
- Valverde S, Cabezas M, Roura E, González-Villà S, Pareto D, Vilanova JC, et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. Neuroimage. 2017;155: 159–168.
- Aslani S, Dayan M, Storelli L, Filippi M, Murino V, Rocca MA, et al. Multi-branch convolutional neural network for multiple sclerosis lesion segmentation. Neuroimage. 2019;196: 1–15.
- Kaur A, Kaur L, Singh A. State-of-the-art segmentation techniques and future directions for multiple sclerosis brain lesions. Arch Comput Methods Eng. 2021;28: 951–977.
- Walsh R, Meurée C, Kerbrat A, Masson A, Hussein BR, Gaubert M, et al. Expert variability and deep learning performance in spinal cord lesion segmentation for multiple sclerosis patients. 2023 IEEE 36th International Symposium on Computer-Based Medical Systems (CBMS). doi:10.1109/cbms58004.2023.00263
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18: 203–211.
- Leener BD, Lévy S, Dupont SM, Fonov VS, Stikov N, Collins DL, et al. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. NeuroImage. 2017;145, Part A: 24–43.